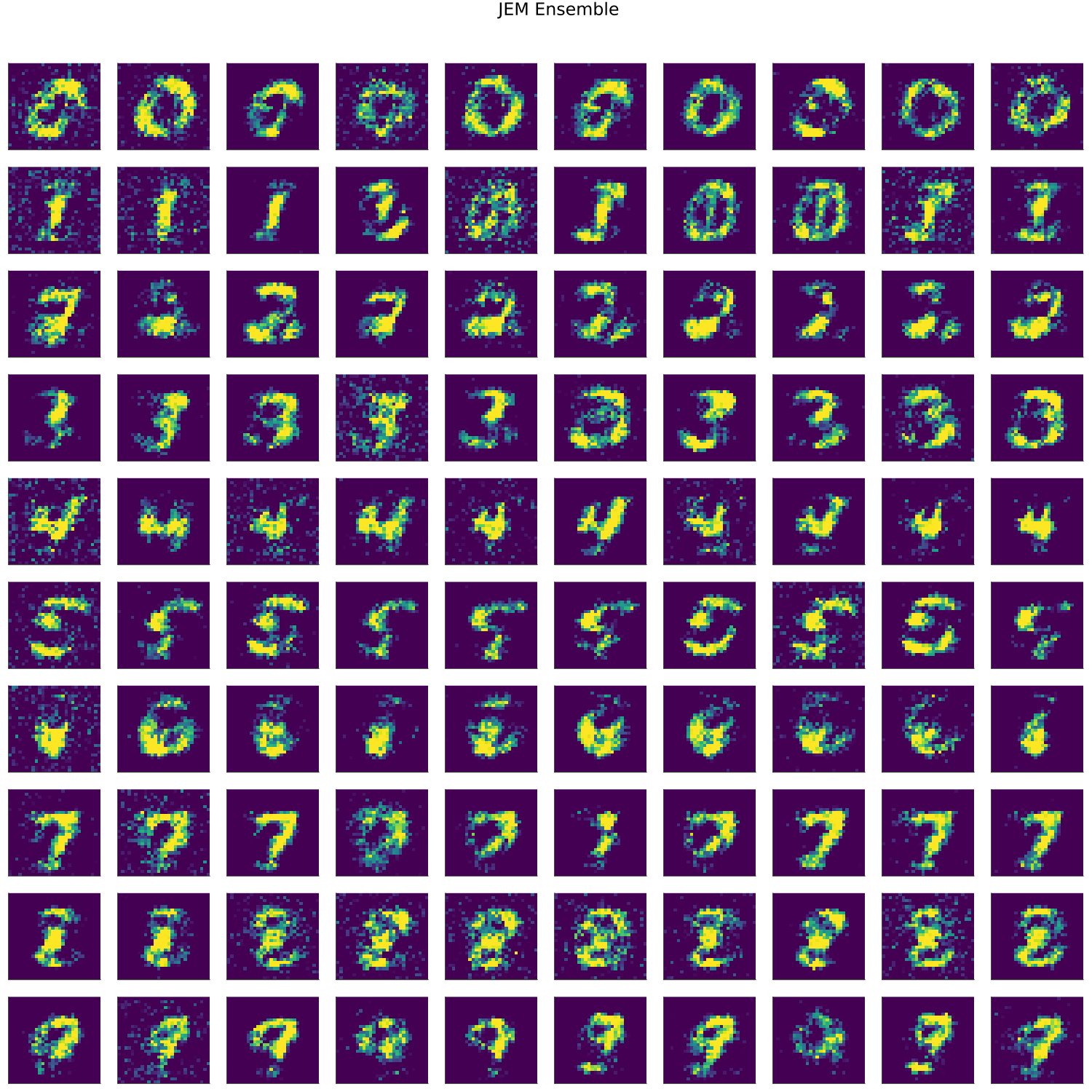
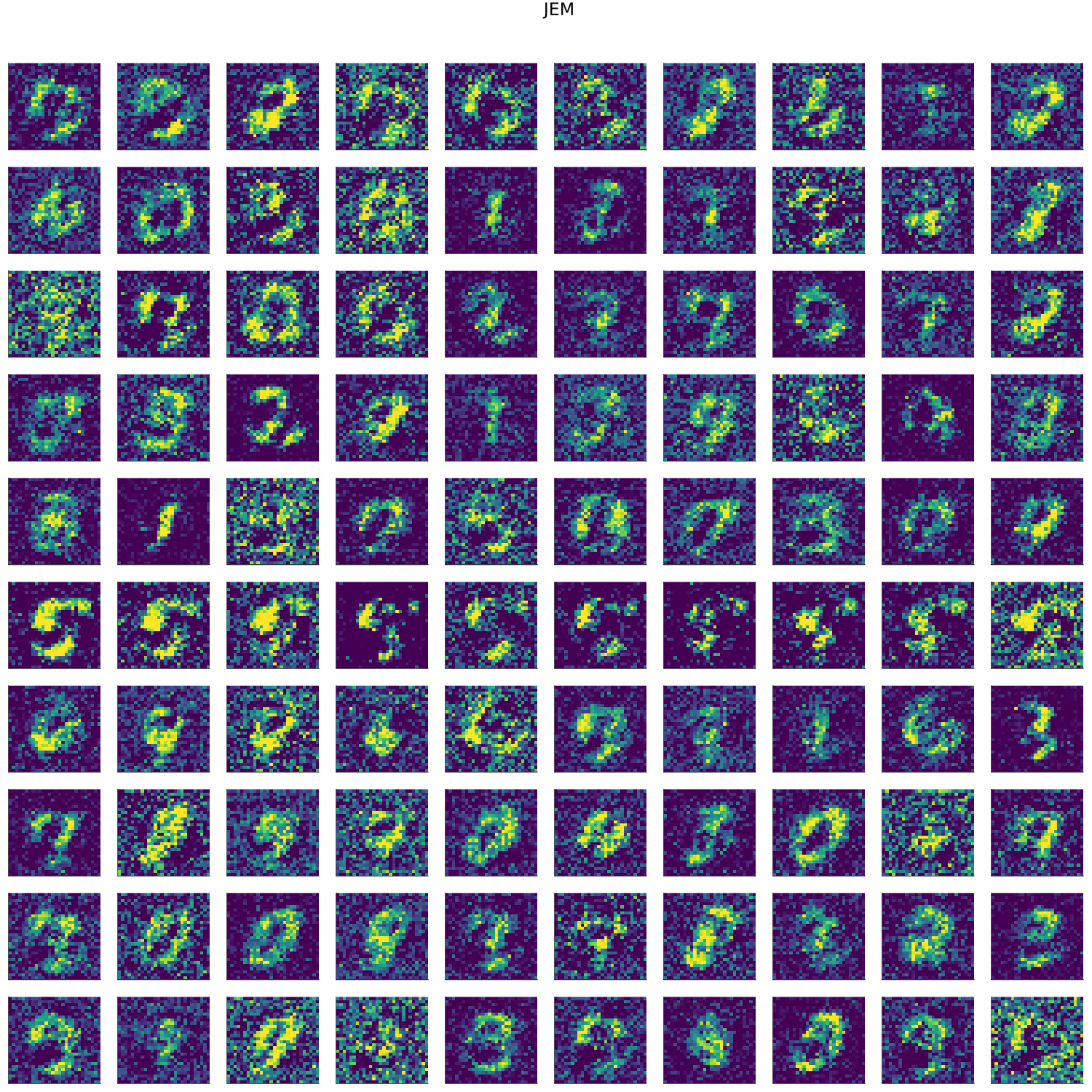
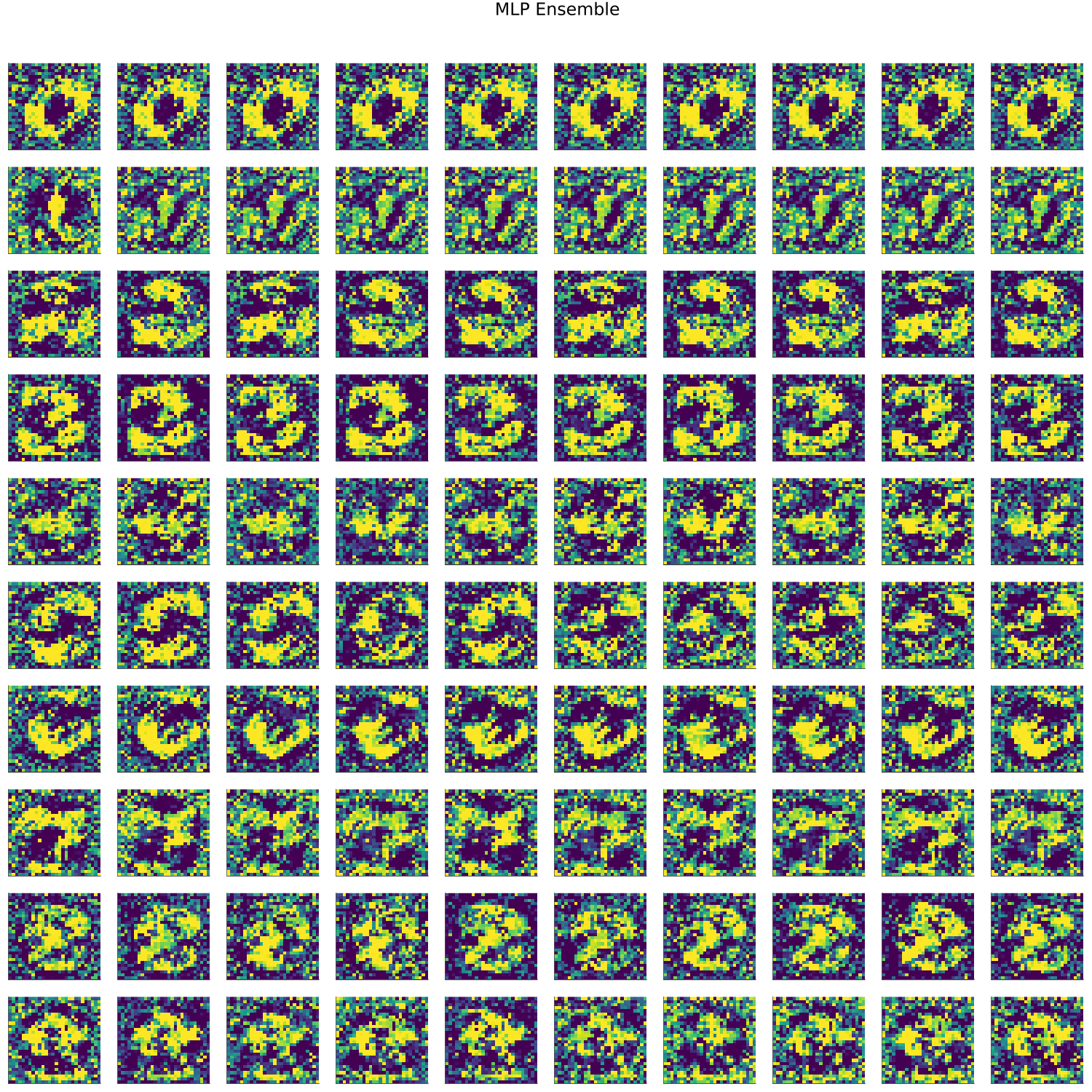
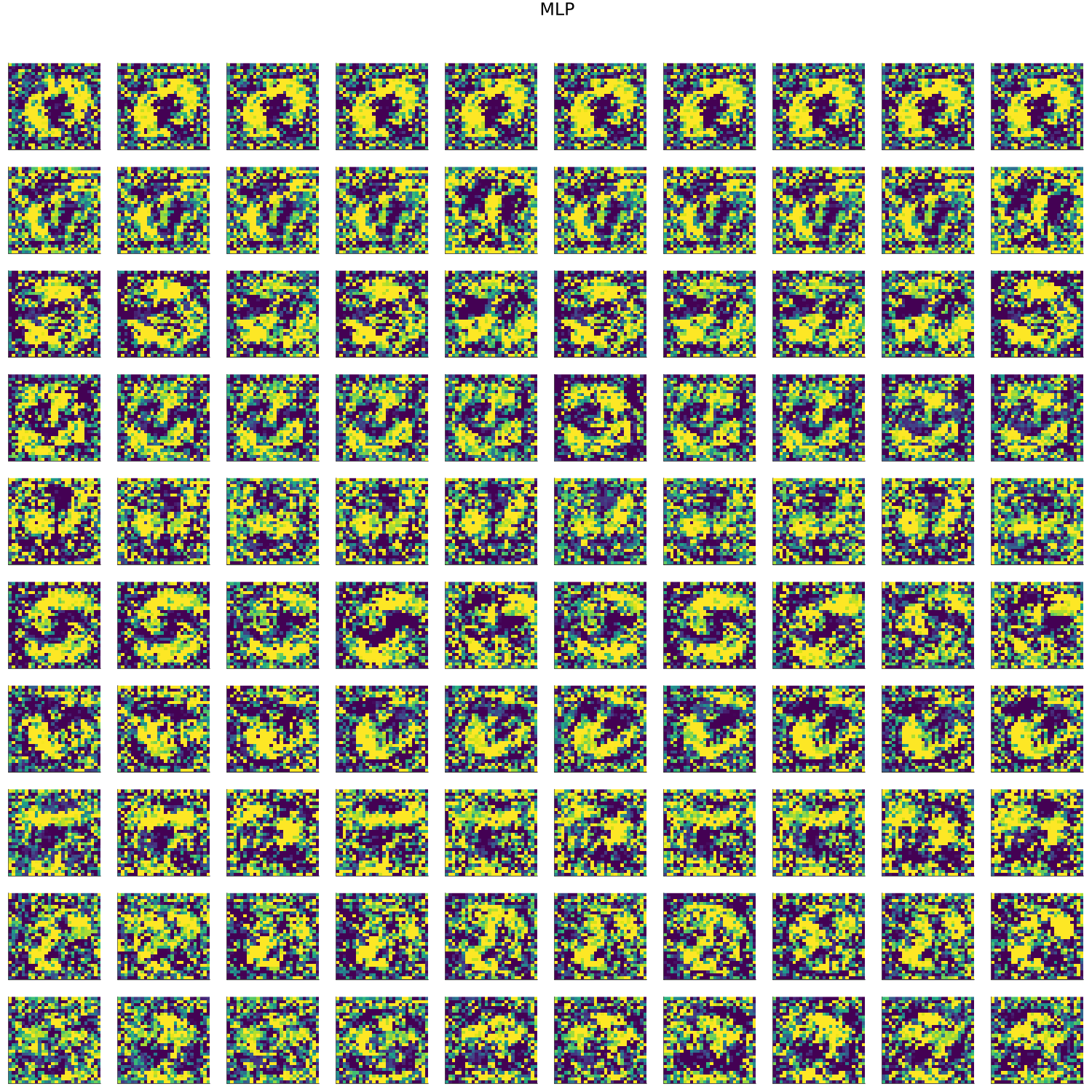
MNIST
Showing
- artifacts/results/images/mnist_generated_JEM Ensemble.png 0 additions, 0 deletionsartifacts/results/images/mnist_generated_JEM Ensemble.png
- artifacts/results/images/mnist_generated_JEM.png 0 additions, 0 deletionsartifacts/results/images/mnist_generated_JEM.png
- artifacts/results/images/mnist_generated_MLP Ensemble.png 0 additions, 0 deletionsartifacts/results/images/mnist_generated_MLP Ensemble.png
- artifacts/results/images/mnist_generated_MLP.png 0 additions, 0 deletionsartifacts/results/images/mnist_generated_MLP.png
- artifacts/results/mnist_model_performance.csv 5 additions, 0 deletionsartifacts/results/mnist_model_performance.csv
- artifacts/results/mnist_model_performance.jls 0 additions, 0 deletionsartifacts/results/mnist_model_performance.jls
- artifacts/results/mnist_models.jls 0 additions, 0 deletionsartifacts/results/mnist_models.jls
- artifacts/results/mnist_vae.jls 0 additions, 0 deletionsartifacts/results/mnist_vae.jls
- artifacts/results/mnist_vae_weak.jls 0 additions, 0 deletionsartifacts/results/mnist_vae_weak.jls
- notebooks/mnist.qmd 83 additions, 9 deletionsnotebooks/mnist.qmd
- notebooks/setup.jl 1 addition, 0 deletionsnotebooks/setup.jl
- paper/paper.pdf 0 additions, 0 deletionspaper/paper.pdf
- paper/paper.tex 2 additions, 1 deletionpaper/paper.tex
- src/generator.jl 1 addition, 1 deletionsrc/generator.jl
- src/model.jl 27 additions, 0 deletionssrc/model.jl
- src/penalties.jl 0 additions, 1 deletionsrc/penalties.jl
- src/sampling.jl 11 additions, 5 deletionssrc/sampling.jl
159 KiB
321 KiB
335 KiB
325 KiB
File added
No preview for this file type
No preview for this file type
No preview for this file type
No preview for this file type